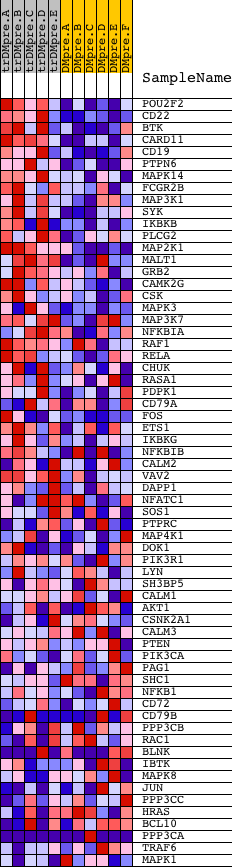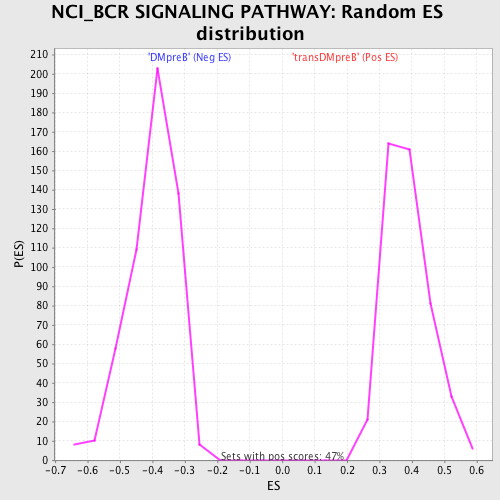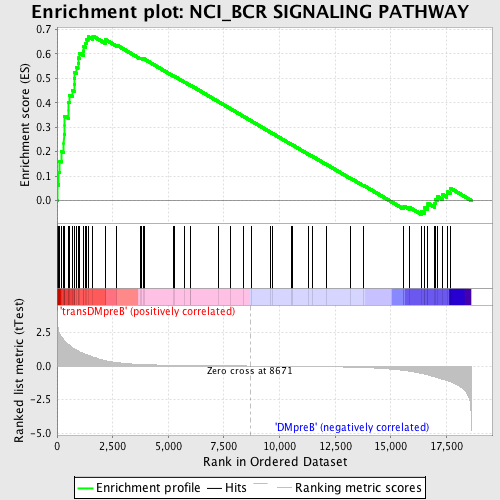
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB.phenotype_transDMpreB_versus_DMpreB.cls #transDMpreB_versus_DMpreB_repos |
| Phenotype | phenotype_transDMpreB_versus_DMpreB.cls#transDMpreB_versus_DMpreB_repos |
| Upregulated in class | transDMpreB |
| GeneSet | NCI_BCR SIGNALING PATHWAY |
| Enrichment Score (ES) | 0.67307526 |
| Normalized Enrichment Score (NES) | 1.738617 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.11429105 |
| FWER p-Value | 0.152 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | POU2F2 | 17929 | 12 | 3.292 | 0.0658 | Yes | ||
| 2 | CD22 | 17881 | 70 | 2.621 | 0.1157 | Yes | ||
| 3 | BTK | 24061 | 118 | 2.378 | 0.1612 | Yes | ||
| 4 | CARD11 | 8436 | 186 | 2.186 | 0.2018 | Yes | ||
| 5 | CD19 | 17640 | 306 | 1.909 | 0.2340 | Yes | ||
| 6 | PTPN6 | 17002 | 309 | 1.905 | 0.2723 | Yes | ||
| 7 | MAPK14 | 23313 | 334 | 1.857 | 0.3086 | Yes | ||
| 8 | FCGR2B | 8959 | 336 | 1.855 | 0.3460 | Yes | ||
| 9 | MAP3K1 | 21348 | 512 | 1.620 | 0.3693 | Yes | ||
| 10 | SYK | 21636 | 513 | 1.619 | 0.4020 | Yes | ||
| 11 | IKBKB | 4907 | 558 | 1.550 | 0.4309 | Yes | ||
| 12 | PLCG2 | 18453 | 681 | 1.392 | 0.4525 | Yes | ||
| 13 | MAP2K1 | 19082 | 767 | 1.285 | 0.4738 | Yes | ||
| 14 | MALT1 | 6274 | 775 | 1.279 | 0.4993 | Yes | ||
| 15 | GRB2 | 20149 | 792 | 1.266 | 0.5240 | Yes | ||
| 16 | CAMK2G | 21905 | 866 | 1.191 | 0.5442 | Yes | ||
| 17 | CSK | 8805 | 940 | 1.117 | 0.5628 | Yes | ||
| 18 | MAPK3 | 6458 11170 | 961 | 1.090 | 0.5837 | Yes | ||
| 19 | MAP3K7 | 16255 | 1018 | 1.035 | 0.6016 | Yes | ||
| 20 | NFKBIA | 21065 | 1178 | 0.947 | 0.6122 | Yes | ||
| 21 | RAF1 | 17035 | 1186 | 0.942 | 0.6308 | Yes | ||
| 22 | RELA | 23783 | 1277 | 0.876 | 0.6437 | Yes | ||
| 23 | CHUK | 23665 | 1310 | 0.853 | 0.6592 | Yes | ||
| 24 | RASA1 | 10174 | 1390 | 0.808 | 0.6712 | Yes | ||
| 25 | PDPK1 | 23097 | 1610 | 0.675 | 0.6731 | Yes | ||
| 26 | CD79A | 18342 | 2161 | 0.395 | 0.6514 | No | ||
| 27 | FOS | 21202 | 2168 | 0.392 | 0.6590 | No | ||
| 28 | ETS1 | 10715 6230 3135 | 2680 | 0.253 | 0.6366 | No | ||
| 29 | IKBKG | 2570 2562 4908 | 3736 | 0.119 | 0.5821 | No | ||
| 30 | NFKBIB | 17906 | 3771 | 0.117 | 0.5826 | No | ||
| 31 | CALM2 | 8681 | 3860 | 0.110 | 0.5801 | No | ||
| 32 | VAV2 | 5848 2670 | 3906 | 0.108 | 0.5799 | No | ||
| 33 | DAPP1 | 11159 6445 6446 | 5222 | 0.053 | 0.5101 | No | ||
| 34 | NFATC1 | 23398 1999 5167 9455 1985 1957 | 5266 | 0.052 | 0.5088 | No | ||
| 35 | SOS1 | 5476 | 5727 | 0.041 | 0.4848 | No | ||
| 36 | PTPRC | 5327 9662 | 5973 | 0.036 | 0.4723 | No | ||
| 37 | MAP4K1 | 18313 | 6010 | 0.036 | 0.4711 | No | ||
| 38 | DOK1 | 17104 1018 1177 | 7255 | 0.017 | 0.4044 | No | ||
| 39 | PIK3R1 | 3170 | 7780 | 0.010 | 0.3764 | No | ||
| 40 | LYN | 16281 | 8372 | 0.004 | 0.3446 | No | ||
| 41 | SH3BP5 | 6278 | 8721 | -0.001 | 0.3258 | No | ||
| 42 | CALM1 | 21184 | 9606 | -0.012 | 0.2784 | No | ||
| 43 | AKT1 | 8568 | 9662 | -0.013 | 0.2757 | No | ||
| 44 | CSNK2A1 | 14797 | 10544 | -0.024 | 0.2287 | No | ||
| 45 | CALM3 | 8682 | 10585 | -0.025 | 0.2271 | No | ||
| 46 | PTEN | 5305 | 11310 | -0.036 | 0.1888 | No | ||
| 47 | PIK3CA | 9562 | 11493 | -0.039 | 0.1798 | No | ||
| 48 | PAG1 | 15376 | 12127 | -0.052 | 0.1467 | No | ||
| 49 | SHC1 | 9813 9812 5430 | 13187 | -0.085 | 0.0913 | No | ||
| 50 | NFKB1 | 15160 | 13771 | -0.113 | 0.0622 | No | ||
| 51 | CD72 | 8718 | 15559 | -0.322 | -0.0276 | No | ||
| 52 | CD79B | 20185 1309 | 15588 | -0.328 | -0.0225 | No | ||
| 53 | PPP3CB | 5285 | 15826 | -0.383 | -0.0275 | No | ||
| 54 | RAC1 | 16302 | 16377 | -0.547 | -0.0461 | No | ||
| 55 | BLNK | 23681 3691 | 16514 | -0.592 | -0.0415 | No | ||
| 56 | IBTK | 19047 | 16518 | -0.594 | -0.0296 | No | ||
| 57 | MAPK8 | 6459 | 16644 | -0.646 | -0.0233 | No | ||
| 58 | JUN | 15832 | 16667 | -0.656 | -0.0113 | No | ||
| 59 | PPP3CC | 21763 | 16969 | -0.809 | -0.0111 | No | ||
| 60 | HRAS | 4868 | 17006 | -0.832 | 0.0037 | No | ||
| 61 | BCL10 | 15397 | 17084 | -0.870 | 0.0172 | No | ||
| 62 | PPP3CA | 1863 5284 | 17331 | -0.993 | 0.0240 | No | ||
| 63 | TRAF6 | 5797 14940 | 17525 | -1.076 | 0.0353 | No | ||
| 64 | MAPK1 | 1642 11167 | 17667 | -1.160 | 0.0512 | No |